2017-上半年-书单
2017-06-29 读书-杂谈 books- 心若菩提

- 失控

- 吃货的生物学修养

- 智能时代:大数据与智能革命重新定义未来

- 批判性思维工具

- 1984

- 中国大历史

- 发条橙

- 计算机与人脑

- 定位:争夺用户心智的战争

- 理性动物

- 创新公司:皮克斯的启示

- DOOM启示录

- 大脑使用手册















A curated list of awesome deep learning applications in the field of neurological image analysis
| Date | Title | Authors | Journal |
|---|---|---|---|
| 2013-06 | Natural Image Bases to Represent Neuroimaging Data | Ashish Gupta, Murat Seckin Ayhan, Anthony S. Maida | Proceedings of the 30th International Conference on Machine Learning (PMLR) |
| 2013-09 | Manifold learning of brain MRIs by deep learning | Brosch, Tom, Roger Tam, and Alzheimer’s Disease Neuroimaging Initiative | 2013 International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) |
| 2013-09 | Unsupervised deep feature learning for deformable registration of MR brain images | Wu, Guorong, Minjeong Kim, Qian Wang, Yaozong Gao, Shu Liao, and Dinggang Shen | 2013 International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) |
| 2013-09 | Unsupervised Deep Learning for Hippocampus Segmentation in 7.0 Tesla MR Images | Kim, Minjeong, Guorong Wu, and Dinggang Shen | 2013 International Workshop on Machine Learning in Medical Imaging |
| 2014-05 | High-level feature based PET image retrieval with deep learning architecture | Liu, Siqi, Sidong Liu, Weidong Cai, Hangyu Che, Sonia Pujol, Ron Kikinis, Michael Fulham, and Dagan Feng | Journal of Nuclear Medicine |
| 2014-08 | Deep learning for neuroimaging: a validation study | Plis, Sergey M., Devon R. Hjelm, Ruslan Salakhutdinov, Elena A. Allen, Henry J. Bockholt, Jeffrey D. Long, Hans J. Johnson, Jane S. Paulsen, Jessica A. Turner, and Vince D. Calhoun | Frontiers in Neuroscience |
| 2014-08 | Restricted Boltzmann Machines for Neuroimaging: an Application in Identifying Intrinsic Networks | Hjelm, R. Devon, Vince D. Calhoun, Ruslan Salakhutdinov, Elena A. Allen, Tulay Adali, and Sergey M. Plis | NeuroImage |
| 2014-09 | Modeling the variability in brain morphology and lesion distribution in multiple sclerosis by deep learning | Brosch, Tom, Youngjin Yoo, David KB Li, Anthony Traboulsee, and Roger Tam | 2014 International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) |
| 2014-09 | Deep learning of image features from unlabeled data for multiple sclerosis lesion segmentation | Yoo, Youngjin, Tom Brosch, Anthony Traboulsee, David KB Li, and Roger Tam | 2014 International Workshop on Machine Learning in Medical Imaging |
| 2014-09 | Segmenting Hippocampus from Infant Brains by Sparse Patch Matching with Deep-Learned Features | Guo, Yanrong, Guorong Wu, Leah A. Commander, Stephanie Szary, Valerie Jewells, Weili Lin, and Dinggang Shen | 2014 International Conference on Medical Image Computing and Computer-Assisted Intervention |
| 2014-09 | Deep Learning Based Imaging Data Completion for Improved Brain Disease Diagnosis | Rongjian Li, Wenlu Zhang, Heung-Il Suk, Li Wang, Jiang Li, Dinggang Shen, Shuiwang Ji | Medical Image Computing and Computer-Assisted Intervention (MICCAI 2014) |
| 2014-10 | Deep learning for brain decoding | Firat, Orhan, Like Oztekin, and Fatos T. Yarman Vural | IEEE International Conference on Image Processing (ICIP) |
| 2014-11 | Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis | Suk, Heung-Il, Seong-Whan Lee, Dinggang Shen, and Alzheimer’s Disease Neuroimaging Initiative | NeuroImage |
| 2014-11 | Multimodal Neuroimaging Feature Learning for Multiclass Diagnosis of Alzheimer’s Disease | Liu, Siqi, Sidong Liu, Weidong Cai, Hangyu Che, Sonia Pujol, Ron Kikinis, Dagan Feng, and Michael J. Fulham | IEEE Transactions on Biomedical Engineering |
| 2014-11 | Classification on ADHD with Deep Learning | Kuang, Deping, and Lianghua He | 2014 International Conference on Cloud Computing and Big Data (CCBD) |
| 2014-12 | Learning Deep Temporal Representations for Brain Decoding | Firat, Orhan, Emre Aksan, Ilke Oztekin, and Fatos T. Yarman Vural | arXiv |
| 2015-01 | Deep learning of fMRI big data: a novel approach to subject-transfer decoding | Koyamada, Sotetsu, Yumi Shikauchi, Ken Nakae, Masanori Koyama, and Shin Ishii | arXiv |
| 2015-02 | Multi-Phase Feature Representation Learning for Neurodegenerative Disease Diagnosis | Liu, Siqi, Sidong Liu, Weidong Cai, Sonia Pujol, Ron Kikinis, and David Dagan Feng | 2015 Australasian Conference on Artificial Life and Computational Intelligence |
| 2015-02 | Predicting Alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks | Adrien Payan, Giovanni Montana | arXiv |
| 2015-03 | Latent feature representation with stacked auto-encoder for AD/MCI diagnosis | Suk, Heung-Il, Seong-Whan Lee, Dinggang Shen, and Alzheimer’s Disease Neuroimaging Initiative | Brain Structure and Function |
| 2015-03 | Deep convolutional neural networks for multi-modality isointense infant brain image segmentation | Zhang, Wenlu, Rongjian Li, Houtao Deng, Li Wang, Weili Lin, Shuiwang Ji, and Dinggang Shen | NeuroImage |
| 2015-05 | A Robust Deep Model for Improved Classification of AD/MCI Patients | Feng Li, Loc Tran, Kim-Han Thung, Shuiwang Ji, Dinggang Shen, and Jiang Li | IEEE JOURNAL OF BIOMEDICAL AND HEALTH INFORMATICS |
| 2015-06 | Deep neural networks for anatomical brain segmentation | de Brebisson, Alexander, and Giovanni Montana | Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops |
| 2015-09 | Deep independence network analysis of structural brain imaging: A simulation study | Castro, Eduardo, Devon Hjelm, Sergey Plis, Laurent Dinh, Jessica Turner, and Vince Calhoun | 2015 IEEE 25th International Workshop on Machine Learning for Signal Processing (MLSP) |
| 2015-10 | Multi-Scale 3D Convolutional Neural Networks for Lesion Segmentation in Brain MRI | Kamnitsas, Konstantinos, Liang Chen, Christian Ledig, Daniel Rueckert, and Ben Glocker | 2015 Ischemic Stroke Lesion Segmentation Challenge (ISLES) |
| 2015-10 | Marginal Space Deep Learning: Efficient Architecture for Volumetric Image Parsing | Ghesu, Florin C., Bogdan Georgescu, Yefeng Zheng, Joachim Hornegger, and Dorin Comaniciu | 2015 International Conference on Medical Image Computing and Computer-Assisted Intervention |
| 2015-11 | Deep Convolutional Encoder Networks for Multiple Sclerosis Lesion Segmentation | Brosch, Tom, Youngjin Yoo, Lisa YW Tang, David KB Li, Anthony Traboulsee, and Roger Tam | 2015 International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) |
| 2016-01 | Deep neural network with weight sparsity control and pre-training extracts hierarchical features and enhances classification performance: Evidence from whole-brain resting-state functional connectivity patterns of schizophrenia | Kim, Junghoe, Vince D. Calhoun, Eunsoo Shim, and Jong-Hwan Lee | NeuroImage |
| 2016-04 | Efficient Multi-Scale 3D CNN with Fully Connected CRF for Accurate Brain Lesion Segmentation | Kamnitsas, Konstantinos, Christian Ledig, Virginia FJ Newcombe, Joanna P. Simpson, Andrew D. Kane, David K. Menon, Daniel Rueckert, and Ben Glocker | arXiv |
| 2016-04 | Multimodal fusion of brain structural and functional imaging with a deep neural machine translation approach | Amin, Md Faijul, Sergey M. Plis, Eswar Damaraju, Devon Hjelm, KyungHyun Cho, and Vince D. Calhoun | 2016 IEEE Southwest Symposium on Image Analysis and Interpretation (SSIAI) |
| 2016-04 | Task-specific feature extraction and classification of fMRI volumes using a deep neural network initialized with a deep belief network: Evaluation using sensorimotor tasks | Jang, Hojin, Sergey M. Plis, Vince D. Calhoun, and Jong-Hwan Lee | NeuroImage |
| 2016-05 | Deep 3D Convolutional Encoder Networks With Shortcuts for Multiscale Feature Integration Applied to Multiple Sclerosis Lesion Segmentation | Brosch, Tom, Lisa YW Tang, Youngjin Yoo, David KB Li, Anthony Traboulsee, and Roger Tam | IEEE transactions on medical imaging |
| 2016-05 | Marginal Space Deep Learning: Efficient Architecture for Volumetric Image Parsing | Ghesu, Florin C., Edward Krubasik, Bogdan Georgescu, Vivek Singh, Yefeng Zheng, Joachim Hornegger, and Dorin Comaniciu | IEEE transactions on medical imaging |
| 2016-05 | q-Space Deep Learning: Twelve-Fold Shorter and Model-Free Diffusion MRI Scans | Golkov, Vladimir, Alexey Dosovitskiy, Jonathan I. Sperl, Marion I. Menzel, Michael Czisch, Philipp Sämann, Thomas Brox, and Daniel Cremers | IEEE transactions on medical imaging |
| 2016-05 | Automatic Detection of Cerebral Microbleeds From MR Images via 3D Convolutional Neural Networks | Dou, Qi, Hao Chen, Lequan Yu, Lei Zhao, Jing Qin, Defeng Wang, Vincent CT Mok, Lin Shi, and Pheng-Ann Heng | IEEE transactions on medical imaging |
| 2016-05 | Automatic Segmentation of MR Brain Images With a Convolutional Neural Network | Moeskops, Pim, Max A. Viergever, Adriënne M. Mendrik, Linda S. de Vries, Manon JNL Benders, and Ivana Išgum | IEEE transactions on medical imaging |
| 2016-05 | Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images | Pereira, Sérgio, Adriano Pinto, Victor Alves, and Carlos A. Silva | IEEE transactions on medical imaging |
| 2016-05 | Clinical decision support for Alzheimer’s disease based on deep learning and brain network | Chenhui Hu, Ronghui Ju, Yusong Shen, Pan Zhou, Li., Q | 2016 IEEE International Conference on Communications (ICC) |
| 2016-06 | Longitudinal Brain Structure Changes in Healthy/Mci Patients: A Deep Learning Approach for the Diagnosis and Prognosis of Alzheimer’s Disease | Peng Dai, Femida Gwadry-Sridhar, Michael Bauer, Michael Borrie, Xue Teng | Alzheimer’s & Dementia |
| 2016-07 | Deep Independence Network Analysis of Structural Brain Imaging: Application to Schizophrenia | Castro, Eduardo, R. Devon Hjelm, Sergey Plis, Laurent Dihn, Jessica Turner, and Vince Calhoun | IEEE Transactions on Medical Imaging |
| 2016-07 | Alzheimer’s Disease Diagnostics by Adaptation of 3D Convolutional Network | Ehsan Hosseini-Asl, Robert Keynto, Ayman El-Baz | 2016 IEEE International Conference on Image Processing |
| 2016-08 | VoxResNet: Deep Voxelwise Residual Networks for Volumetric Brain Segmentation | Chen, Hao, Qi Dou, Lequan Yu, and Pheng-Ann Heng | arXiv |
| 2016-10 | Deep Learning of Brain Lesion Patterns for Predicting Future Disease Activity in Patients with Early Symptoms of Multiple Sclerosis. | Yoo, Youngjin, Lisa W. Tang, Tom Brosch, David KB Li, Luanne Metz, Anthony Traboulsee, and Roger Tam | Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings |
| 2016-10 | Convolutional Neural Network for Reconstruction of 7T-like Images from 3T MRI Using Appearance and Anatomical Features | Bahrami, Khosro, Feng Shi, Islem Rekik, and Dinggang Shen | Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings |
| 2016-10 | Longitudinal Multiple Sclerosis Lesion Segmentation Using Multi-view Convolutional Neural Networks | Birenbaum, Ariel, and Hayit Greenspan | Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings |
| 2016-10 | De-noising of Contrast-Enhanced MRI Sequences by an Ensemble of Expert Deep Neural Networks | Benou, Ariel, Ronel Veksler, Alon Friedman, and Tammy Riklin Raviv | Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings |
| 2016-10 | Multi-dimensional Gated Recurrent Units for the Segmentation of Biomedical 3D-Data | Andermatt, Simon, Simon Pezold, and Philippe Cattin | Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings |
| 2016-10 | Learning Thermal Process Representations for Intraoperative Analysis of Cortical Perfusion During Ischemic Strokes | Hoffmann, Nico, Edmund Koch, Gerald Steiner, Uwe Petersohn, and Matthias Kirsch | Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings |
| 2016-10 | 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients | Dong Nie, Han Zhang, Ehsan Adeli, Luyan Liu, Dinggang Shen | Medical Image Computing and Computer-Assisted Intervention (MICCAI 2016) |
| 2016-12 | Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker | James H Cole, Rudra PK Poudel, Dimosthenis Tsagkrasoulis, Matthan WA Caan, Claire Steves, Tim D Spector, Giovanni Montana | arXiv |
| 2016-12 | DeepAD: Alzheimer’s Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI | Saman Sarraf, John Anderson, Ghassem Tofighi | bioRxiv |
| 2017-01 | Brain tumor segmentation with Deep Neural Networks | Havaei, Mohammad, Axel Davy, David Warde-Farley, Antoine Biard, Aaron Courville, Yoshua Bengio, Chris Pal, Pierre-Marc Jodoin, and Hugo Larochelle | Medical Image Analysis |
| 2017-02 | Generic decoding of seen and imagined objects using hierarchical visual features | T Horikawa, Y Kamitani | Nat Commun |
| 2017-02 | DeepNAT: Deep convolutional neural network for segmenting neuroanatomy | Christian Wachingera, Martin Reuterb, Tassilo Klein | NeuroImage |
| 2017-03 | Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications | Sandra Vieira, Walter H.L. Pinaya, Andrea Mechelli | Neuroscience & Biobehavioral Reviews |
| 2017-04 | Predicting Cognitive Decline with Deep Learning of Brain Metabolism and Amyloid Imaging | Hongyoon Choi, Kyong Hwan Jin | arXiv |
| 2017-04 | Deep ensemble learning of sparse regression models for brain disease diagnosis | Heung-Il Suk, Seong-Whan Leea, Dinggang Shen | Medical Image Analysis |
Feel free to send a pull request. Use DOI links, when possible.
特征选择在很多方面都非常有用:它是对抗维数灾难的最好武器;它能减少训练时间;它还是减轻过拟合、提高泛化性能的有利手段。 By Matthew Mayo, KDnuggets
比如你想对动物进行分类,基于一个过剩的数据集,你会很快发现各种可能的数据属性(特征)对分类都没什么用。例如所有活的生物都有且只有一个心脏,这个特征从学习的角度来说是没有用的。从另一方面讲,一个指明当前动物是否有蹄子的特征会是一个很好的预测量变量(predictor)。
进一步讲,将不相关的特征和非常有效的预测变量混合会对预测模型造城非常不好的影响。也就是说,加入无关特征会增加训练时间,并可能导致在训练数据集的过拟合。
特征选择是缩小特征子集的过程,通常用于预测建模。特征选择在很多方面都非常有用:它是对抗维数灾难的最好武器;它能减少训练时间;他还是减轻过拟合、提高泛化性能的有利手段。
我最近读到的数据科学家Rubens Zimbres的文章 – written so eloquently and concisely – 从现实角度提及了特征选择的重要性:
经过实践,使用层叠神经网络,平行神经网络,不对称配置,简单神经网络,multiple layers,dropouts,activation functions等等技术得到一个结论:都不如特征选择好使。
我曾经与Rubens Zimbres进行过专业交流,我请求他传授些技巧,他的分享如下:
特征选择应该成为一个数据科学家主要的关注点之一。特征选择能够提高正确率和泛化性能,藉由相关性、偏度(skewness)、t-test、ANOVA、熵、信息增益等。
许多时候正确的特征选择能使你训练出更加简单更加快速的机器学习模型。请看下面的图片(SVM对IRIS数据集分类): 左侧代表错误的变量选择,右侧袋面正确的变量选择。线性核不能很好地解决分类任务,rbf核也不是很理想。在右侧,花瓣长和花瓣宽这两个属性被选中作为特征,此时线性核就能很好地进行区分。正确地特征选择,合适的算法,高效地调参是成功地关键。
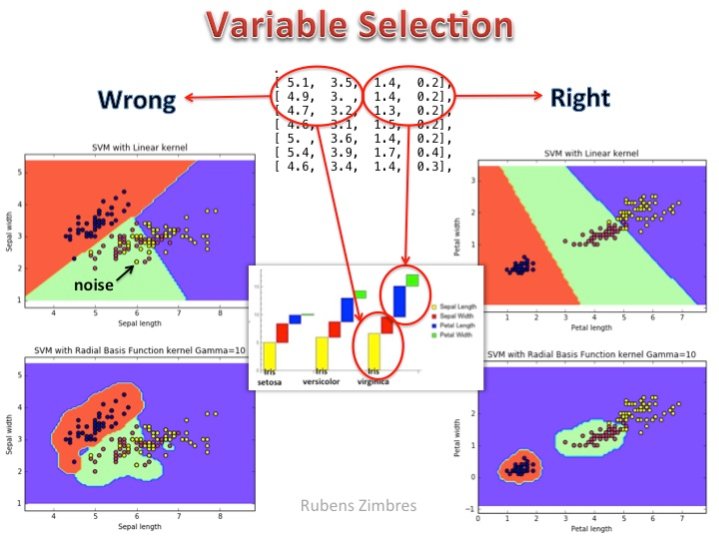
最近以来,大量的数据预处理技术使得我们开始忽视特征选择的意义,但需要记住的是这种情况只适用于特征选择只是用来减少训练时间的应用场合。如Zimbre提到的一个简单而具体的例子,特征选择是区分一个高效的泛化好的模型和一个纯属浪费时间模型的重要标志。
P.S.
推荐两篇神经影像领域应用特征选择算法的论文:
| Date | Title | Authors | Journal |
|---|---|---|---|
| 2013 | A Review of Feature Reduction Techniques in Neuroimaging | Benson Mwangi & Tian Siva Tian & Jair C. Soares | Neuroinform |
| 2016 | Comparison of Feature Selection Techniques in Machine Learning for Anatomical Brain MRI in Dementia | *J Tohka, E Moradi, H Huttunen | Neuroinform |
The Men Who Build the America - 4 episodes (Netease Open Course)

Positive Psychology - 23 lectures (Netease Open Course)

The C++ Programing Language - 6 chapters (TsinghuaX)

Introduction to Computer Science and Programming Using Python - 9 weeks (edX)

Introduction to Computational Thinking and Data Science - 10 weeks (edX)

Computational Neuroscience - 8 weeks (Coursera)
![]()
Data Scientists’ Toolbox - 4 weeks (Coursera)

R Programming - 4 weeks (Coursera)

Introduction to Neurohacking In R - 4 weeks (Coursera)

Learning How to Learn: Powerful mental tools to help you master tough subjects - 4 weeks (Coursera)

Statistical Learning - 10 chapters (Stanford Online)

Neural Networks for Machine Learning - 16 weeks (Coursera)
![]()